VariantDB
UZA-USERS :All data of UZA accounts has been migrated & accounts were disabled here!Use : https://variantdb.uza.be/'
Documentation : Load Samples Into VariantDB
Data can be loaded either by uploading it to an FTP server, or by sending it directly from a galaxy server.
1. FTP upload
- Go to : New Data in the top menu.
- If your system administrator has enabled ftp upload, you should see the instructions to connect to the server. It might look like:

- After uploading VCF/BAM files to FTP server, come back to the New Data page. You will now see a selection box on top containing the uploaded data:
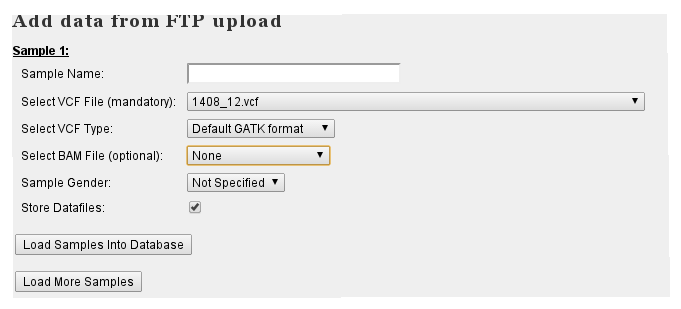
- Provide the requested information and select the data files to store in VariantDB. If 'Store Datafiles' is checked, data can be loaded directly to IGV from VariantDB
- To load more samples at the same time, select 'Load More Samples'
- Select 'Load Samples Into Database'. A status screen is shown. You can safely close this screen
- Upon completion of the import, you will recieve an email notification
2. Import from Galaxy
- Install the VariantDB tool for galaxy. This is available from the toolshed here.
- A publicly available galaxy server with this tool installed is available here
- Make sure you use the EXACT same username in galaxy and VariantDB
- Generate a VCF file (preferably with GATK Unified Genotyper
- Open the 'VCF To VariantDB' tool:
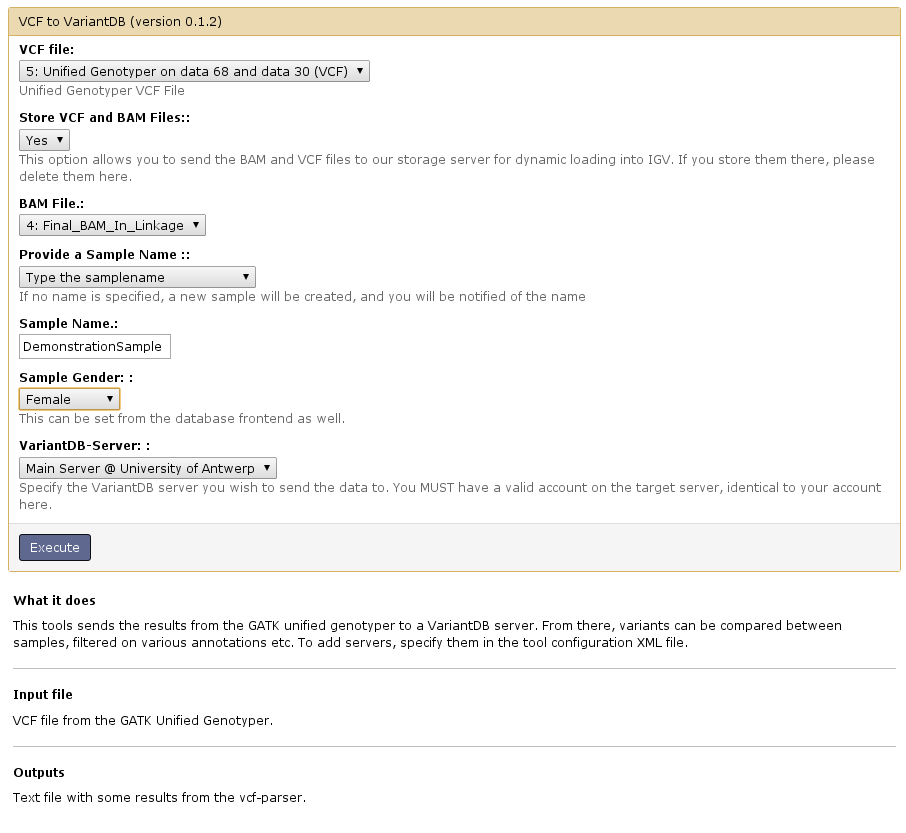
- Provide necessary information and select execute. Import will be finished when the job finishes.